KNOSSOS
Not your system? See all releases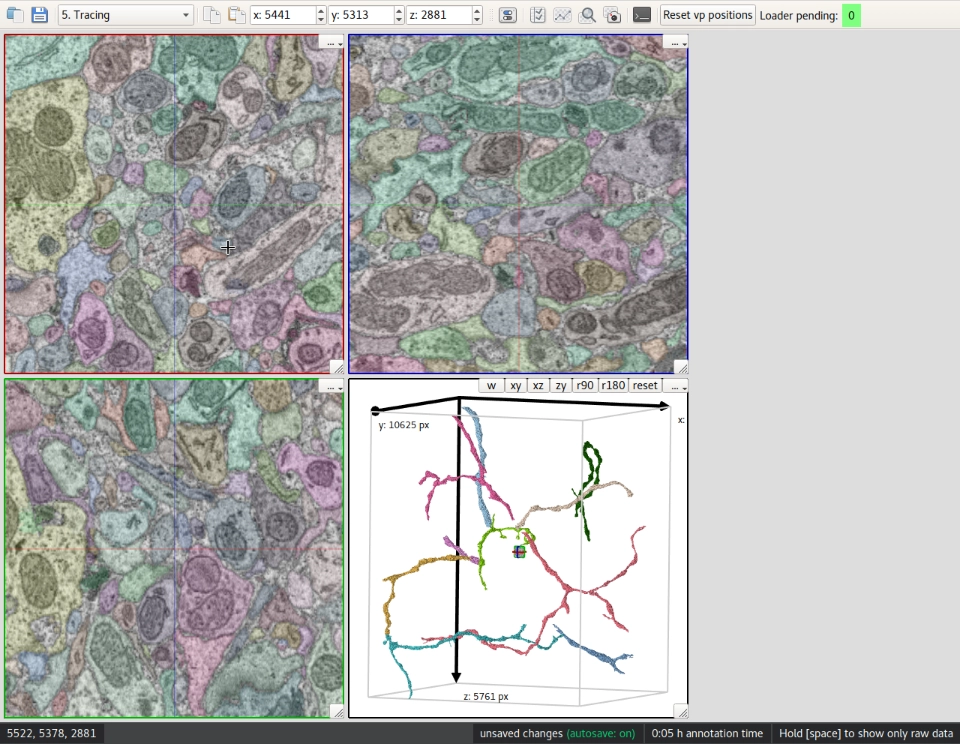

KNOSSOS visualizes large 3D (up to multiple terabyte) volume electron microscopic (e.g. Serial Block-Face EM) datasets by displaying slices. It supports skeleton and volume-based annotation modes, which can be extended by plugins written in Python.
It is developed at the Max Planck Institute for Medical Research in Heidelberg, Germany, for Windows, GNU/Linux and OS X. A similar, web-based implementation is being developed at webknossos.org.
Take a look at KNOSSOS’ features to learn more about it.
3D visualization of image datasets is done by displaying a 2D representation of each side, and allowing you to navigate through these image planes. By dynamically loading only data from the surrounding of the current location, seamless navigation is not limited to datasets that fit into the available RAM but also works with much larger datasets stored in KNOSSOS’ special format on disk.
KNOSSOS supports two annotation methods—skeletonization as well as 3D segmentation for volume reconstruction. Skeletonization is done by placing and connecting nodes, while KNOSSOS’ segmentation mode allows manual processing of pre-segmented data, and creation of new segmentations from scratch.
These features are already being used at Max Planck Institute for Medical Research (among others), where mice retina was successfully reconstructed.
You can enhance KNOSSOS’ annotation features by writing Python plugins, and we also provide a Python script that helps you to convert your existing image data into a KNOSSOS-readable format.
KNOSSOS is developed using the Qt5 toolkit, and available on all major platforms. You can help the development of KNOSSOS by submitting bugs and other suggestions at Github’s issue tracker or by contacting us directly.
| Date of Publication | Title of Publication | DOI Link |
|---|---|---|
| Cell type specific labeling and partial connectomes of dopaminergic circuits reveal non-synaptic communication and large-scale axonal remodeling after exposure to cocaine | https://doi.org/10.1101/2020.09.29.318881 | |
| Permeabilization-free en bloc immunohistochemistry for correlative microscopy | https://doi.org/10.7554/eLife.63392 | |
| Multi-modal imaging of a single postmortem mouse brain over five orders of magnitude of resolution | https://doi.org/10.1016/j.neuroimage.2021.118250 | |
| Primate neuronal connections are sparse as compared to mouse | https://doi.org/10.1101/2020.09.24.311852 | |
| Three-dimensional Fib-Sem reconstruction of microtubule-organelle interaction in whole primary mouse beta cells | https://doi.org/10.1083/jcb.202010039 | |
| Synapse-specific direction selectivity in retinal bipolar cell axon terminals | https://doi.org/10.1101/2020.10.12.335810 | |
| Imaging Plant Cells by High-Pressure Freezing and Serial block-face scanning electron microscopy | https://doi.org/10.1007/978-1-0716-0767-1_7 | |
| Image Processing for Volume Electron Microscopy | https://doi.org/10.1007/978-1-0716-0691-9_13 | |
| Learning cellular morphology with neural networks | https://doi.org/10.1038/s41467-019-10836-3 | |
| EM connectomics reveals axonal target variation in a sequence-generating network | https://doi.org/10.7554/eLife.24364 | |
| Automated synaptic connectivity inference for volume electron microscopy | https://doi.org/10.1038/nmeth.4206 | |
| Volume Electron Microscopic Analyses in the Larval Zebrafish | https://doi.org/10.11588/heidok.00022556 | |
| When complex neuronal structures may not matter | https://doi.org/10.7554/eLife.23508 | |
| Connectivity map of bipolar cells and photoreceptors in the mouse retina | https://doi.org/10.7554/eLife.20041 | |
| 3-dimensional electron microscopic imaging of the zebrafish olfactory bulb and dense reconstruction of neurons | https://doi.org/10.1038/sdata.2016.100 | |
| Species-specific wiring for direction selectivity in the mammalian retina | https://doi.org/10.1038/nature18609 | |
| Dense EM-based reconstruction of the interglomerular projectome in the zebrafish olfactory bulb | https://doi.org/10.1038/nn.4290 | |
| Connectomic reconstruction of the inner plexiform layer in the mouse retina | https://doi.org/10.1038/nature12346 | |
| High-accuracy neurite reconstruction for high-throughput neuroanatomy | https://doi.org/10.1038/nn.2868 | |
| Wiring specificity in the direction-selectivity circuit of the retina | https://doi.org/10.1038/nature09818 |
Your paper? If you think KNOSSOS might be useful for your research, feel free to contact us. We are happy to exchange ideas and to provide assistance!
A special image format is required to use KNOSSOS. You have the choice to either try our example datasets, or learn how to create your own.
The built-in datasets are called e2006, ek0563, and j0256.
An external dataset can be loaded into KNOSSOS by selecting its .conf file in , and clicking on Use.
If you have your own image datasets, they will probably need to be converted into KNOSSOS’ format. Take a look at our documentation to learn how to do so.
These are offline datasets that already contain all pre-formatted images:
KNOSSOS is being actively maintained to provide a useful tool to the study of connectomics. If you miss a feature that would make KNOSSOS more useful for your research, please tell us on our issue tracker on GitHub or via e-mail.
If you encounter any bugs while using KNOSSOS, please tell us as well.
KNOSSOS can be integrated into your existing workflow, either by using the main program or its Python plugin interface.
You can contact us if you need help in writing Python plugins for KNOSSOS or if you have an idea how to improve KNOSSOS’ integration with your workflow.
We want to work closely together with other research teams in the connectomics field. Feel free to contact us if you need any technical assistance in this area or if you want to exchange ideas.
KNOSSOS is developed by a global team of open source contributors — get in touch with us if you want to help!

Research Group Leader

CEO, ariadne.ai (Germany) GmbH

M.Sc., Computer Science

Software Engineer, ariadne.ai (Germany) GmbH

B.A., Computational Linguistics
We would like to thank the following persons for their contributions towards KNOSSOS:
Konrad Kühne, Andreas Knecht, Patrick Müller, Michael Pronkin, Claus Ripp, Oren Shatz, Alexander Stepanov, Matthias Wegner
If you have any questions or suggestions regarding KNOSSOS, feel free to write us: